A Microscopists' View of Chromosome Organization

The purpose of this presentation is to show how cytologists and cytochemists contribute to our understanding of the organization of nuclear chromatin. Chromatin is the name that describes nuclear material that contains the genetic code. In fact, the code is stored in individual units called "chromosomes". As we shall see below, these are highly ordered, organized packages designed for the storage of genetic material, its condensation during cell division and regulation of gene expression.
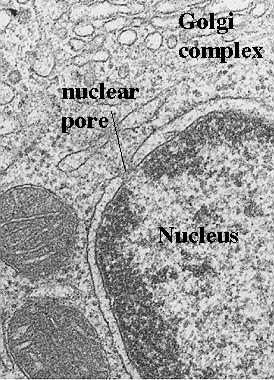
If one uses classical transmission electron microscopy (on glutaraldehyde and osmium-tetroxide-fixed tissues and ultrathin plastic sections), there is no hint of the high degree of organization actually found in the nucleus. When one looks at the nucleus, one can find some important clues about the physiological state of the cell. The drawing in this figure shows the two basic cytological patterns that are evident with classical techniques. Two types of chromatin can be described as follows:

Heterochromatin
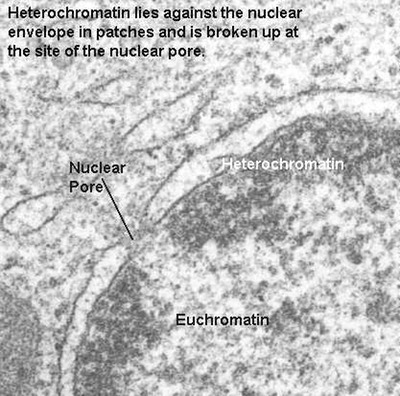
This is the condensed form of chromatin organization. It is seen as dense patches of chromatin. Sometimes it lines the nuclear membrane, however, it is broken by clear areas at the pores so that transport is allowed. Sometimes, the heterochromatin forms a "cartwheel" pattern. Abundant heterochromatin is seen in resting, or reserve cells such as small lymphocytes (memory cells) waiting for exposure to a foreign antigen. Heterochromatin is considered transcriptionally inactive.
See Alberts et al, Molecular Biology of the Cell, Garland Publishing, 1994, pages 352 and 353
Euchromatin
Euchromatin is threadlike, delicate. It is most abundant in active, transcribing cells.
(See Alberts et al, Molecular Biology of the Cell, Garland Publishing, 1994, pp 351-352).
Thus, the presence of euchromatin is significant because the regions of DNA to be
transcribed or duplicated must uncoil before the genetic code can be read.
Return to Menu
Structure of isolated metaphase chromosomes

Just before cell division (and after DNA synthesis), the chromatin condenses further into individual metaphase chromosomes. The dividing chromosomes appear as two chromatids. An electron microscopic view of a chromosome is shown in this photograph. The chromatids appear to be made of coiled loops of 20-30 nm nucleoprotein fibers.
Modified from Bloom and Fawcett, A Textbook of Histology, Chapman and Hall, 12th edition, Figure 1-14
Can one define individual genes on chromosomes?
In the section below, we will show the electron microscopic views that support the
organization of DNA in chromosomes. First, we will look at how we can identify specific
genes by cytochemistry.
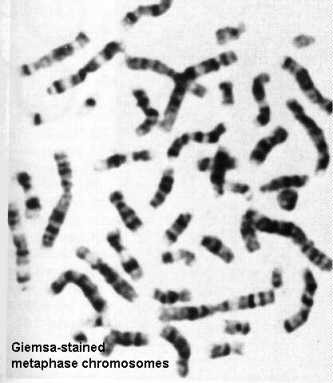
As is evident from the foregoing figure, it is difficult to appreciate details of chromosome structure with the electron microscope. However, one can spread these chromosomes on a light microscope slide and label them with dyes that are preferentially taken up by certain regions (for example, modified Giemsa stains). These modifications create a banding pattern that can be used to identify and characterize individual chromosomes. The banding pattern can be used along with shape and length of arms to identify genetic translocations.
See also Alberts et al, Molecular Biology of the Cell, Garland Pub., N.Y., 1994 pp 355 and 356.
FISH
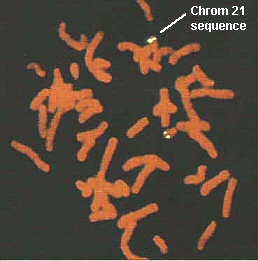
A more precise way to identify genetic sequences and genes is via a technique called "Fluorescence In situ Hybridization" or FISH. This involves the use of DNA or oligonucleotides complementary to a nucleotide sequence on the chromosomes. These "cDNA probes" are conjugated to fluorescein, rhodamine, Texas red, or another fluorescent compound and then used to detect the sites of the gene on the individual chromosomes. This figure detects a sequence on chromosome 21 in a normal individual (white label). Note, there are two copies of this chromosome, each bearing the sequence. This probe is useful for the detection of Down syndrome which contains an extra chromosome 21.
Taken from Boehringer Mannheim Biochemicals "Non Radioactive In Situ Hybridization Application Manual", Figure 10.

FISH has also been called "chromosome painting" and there are commercial kits available to detect multiple chromosomes in one section or metaphase spread. A recently developed kit is so colorful, it has been named "Joseph's coat". Each nucleotide probe is attached to a different fluorescent compound and they can be added together because they will attach to different chromosomes, or different regions of the nucleus or chromosomes. In this photograph, FISH was used to detect sequences in chromatin in interphase nuclei. A triple labeling protocol was applied to detect chromosome 2 (red; Texas red), 4 (blue, AMCA) and 8 (green, fluorescein).
Taken from Boehringer Mannheim Biochemicals "Non Radioactive In Situ Hybridization Application Manual", Figure 7.
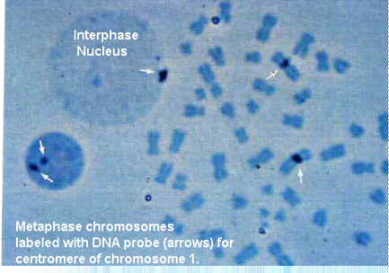
Actually, chromosome painting may involve other types of labeling tools, such as autoradiography for radioactive probes, or immunocytochemistry for biotin-labeled or digoxygenin-labeled probes. The immunocytochemical reaction can be detected with gold, peroxidase or alkaline phosphatase labels. Furthermore, the detection of genes or genetic sequences can be done at the electron microscopic level. This figure shows a metaphase spread next to an interphase nucleus. The centromeric heterochromatin has been detected on chromosome 1 with a specific complementary DNA (cDNA) probe.
Taken from Boehringer Mannheim Biochemicals "Non Radioactive In Situ Hybridization Application Manual", Figure 14.
Return to MenuHow DNA and Histones are organized in chromosomes.
See Alberts et al, Molecular Biology of the Cell, Garland Publishing, 1994, pages 342-346.
To understand how the DNA and histones are organized in a chromosome, we must appreciate the fact that the nucleus is only 6 micrometers in diameter. The total length of DNA in the human genome is 1.8 meters. Thus, in order to pack the DNA into the nucleus as in the photograph of the metaphase chromosome , there must be several levels of coiling and supercoiling. There is nearly a 10,000-fold reduction in length in an interphase nucleus. Each chromosome contains 1 long molecule of DNA plus associated histones (basic proteins) which help in the condensation and regulation processes. In the following section, we will show an uncoiled chromosome and also visualize it with specific electron microscopic techniques.
Visualize 10-30 nm fibers and fibrils
To look at chromatin, we can isolate nuclei and disrupt them. This will disperse the
chromatin which can be spread on a water surface. This spread can be picked up on a
plastic-coated grid and examined with the electron microscope. 
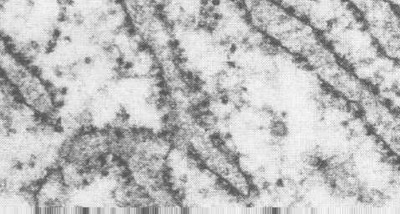
The first level of organization you see is a tangle of 20-30 nm fibers. These are actually coils of the DNA and histones. The figure on the left shows the tangled chromatin fibers in the left panel. Shearing forces can be used to further uncoil and stretch these fibers and the beaded filaments appear. The strands between the beads are segments of double stranded DNA. These are shown in the right panel.
Modified from Bloom and Fawcett, A Textbook of Histology, Chapman and Hall, 12th edition, Figure 1-12
 Visualize 4 nm DNA filaments.
Visualize 4 nm DNA filaments.
Another way to look at the chromatin is to remove the histones from isolated chromatin (with dextran sulfate and heparin). This will cause an unraveling of the chromatin from a central protein core. They can be spread on a water surface and collected with a plastic coated grid for electron microscopy. In the EM, they will appear as 4 nm DNA filaments, with no attached histones. This figure shows the unraveled chromatin in the upper inset with the dense protein core. The individual filaments are seen in the main photograph and in the lower inset at higher magnification.
Modified from Bloom and Fawcett, A Textbook of Histology, Chapman and Hall, 12th edition, Figure 1-15
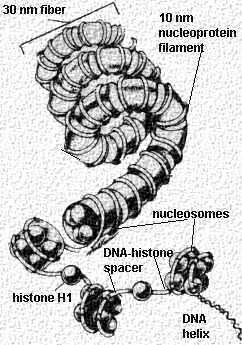 This cartoon
shows the different levels of uncoiling seen in the chromosome. Start from the bottom of
the figure with the DNA helix. Click the hyperlinked text to take you to the electron
micrograph illustrating that level of coiling. The first level is the 4 nm DNA filament seen in the above figure (which is DNA
stripped of its histones). In this cartoon, the 4 nm DNA
filaments are labeled the "DNA helix". In the normal, unstripped chromosome,
the double stranded DNA is wrapped around sets of 8 histones to form a 10 nm filament.
These sets of 8 histones are separated by spacer regions of 4 nm DNA filament (double
stranded DNA) and Histone H1. The sets of 8 histones wrapped by DNA are called
"nucleosomes". These are shown in the cartoon. They are the 10 nm nucleoprotein fibrils or "beads on a string"
seen in the above electron micrographs. These nucleosomes (beads) are separated by the
spacer DNA (seen in the right panel).
This cartoon
shows the different levels of uncoiling seen in the chromosome. Start from the bottom of
the figure with the DNA helix. Click the hyperlinked text to take you to the electron
micrograph illustrating that level of coiling. The first level is the 4 nm DNA filament seen in the above figure (which is DNA
stripped of its histones). In this cartoon, the 4 nm DNA
filaments are labeled the "DNA helix". In the normal, unstripped chromosome,
the double stranded DNA is wrapped around sets of 8 histones to form a 10 nm filament.
These sets of 8 histones are separated by spacer regions of 4 nm DNA filament (double
stranded DNA) and Histone H1. The sets of 8 histones wrapped by DNA are called
"nucleosomes". These are shown in the cartoon. They are the 10 nm nucleoprotein fibrils or "beads on a string"
seen in the above electron micrographs. These nucleosomes (beads) are separated by the
spacer DNA (seen in the right panel).
The next level of coiling produces the 30 nm nucleoprotein fibers . This is the seen in the left panel (above). Further looping of these nucleoprotein fibers around a protein scaffold forms the individual metaphase chromosomes
Modified from Bloom and Fawcett, A Textbook of Histology, Chapman and Hall, 12th edition, Figure 1-13
For more information, contact:
Gwen Childs, Ph.D.,FAAA
Professor and Chair
Department of Neurobiology and Developmental Sciences
University of Arkansas for Medical Sciences
Little Rock, AR 72205
For questions, contact this email address: